Phylogenetic analysis using Machine learning
Introduction:
The interpretation of the phylogenetic tree is an essential yet challenging aspect of evolutionary studies. To conduct an evolutionary study of the organisms is the core of biological research. The resulting phylogeny is then subjected to a plethora of analyses essential for further genomic research (Azouri 2021). The phylogenetic analysis involves several methods that can be used to interpret data. Recently, researchers have begun studying the use of machine learning in inferring phylogenetic trees.

Phylogenetic Analysis:
The study of the evolutionary history of a species or a group of organisms is known as phylogenetic analysis. Here, the evolutionary relationship between different species or organisms having a common ancestor is represented with the help of branching diagrams. This diagram is called the phylogenetic tree, which can be either rooted or unrooted. Phylogenetic analysis can also be used to study the relationship between characteristics of an organism, including genes and proteins.
The applications of phylogenetic analysis are numerous. These include – reconstruction of the ancestral gene for the derivation of extant genes, study of human disease and epidemiology, interpretation of the evolution of ecological and behavioural traits, estimation of historical biogeographic relationships, and many more.
Interesting Blog: Performance Evaluation Metrics for Machine-Learning Based Dissertation
Currently available methods for inference:
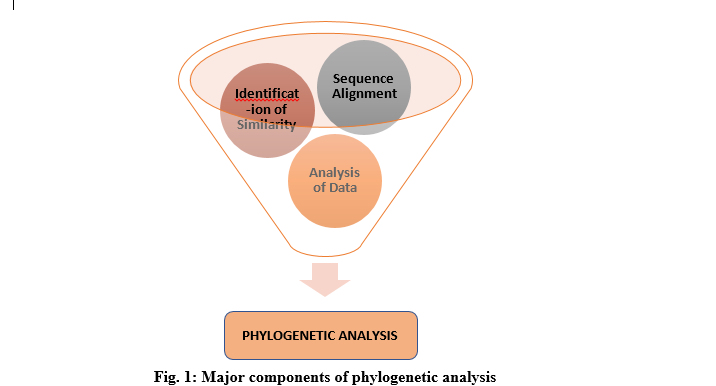
Previously, morphological features were used in the assessment of similarities among species and in phylogenetic analysis. It has drastically changed over time. Nowadays, this analysis uses information extracted from DNA, RNA or protein. The generation of a phylogenetic tree involves the alignment of sequences. The most widely-used tool for this is the alignment-based methodology. In this method, the two sequences are stacked in a way to highlight their common symbols and substrings. This comparison of sequences helps to identify patterns of shared ancestry between species. (Munjal 2019). However, exploiting these large-scale molecular data poses significant challenges. One of the most difficult tasks is to develop effective techniques for the extraction of missing data.
The Maximum likelihood or Markov Chain Monte Carlo (MCMC) methods and probabilistic models of sequence evolution are highly reliable statistical methods used for the reconstruction of gene and species trees. Even so, many of these approaches are not scalable enough to study phylogenomic datasets of hundreds or thousands of genes and taxa. Thus, the development of a quick and efficient method is the need of the hour ( Bhattacharjee 2020).
Application of machine learning:
Machine learning has found various applications in the field of technology-driven research. One such usage of machine learning is in the inference of the phylogenetic tree. In a recent study, researchers utilized the machine learning method to predict the best model for the most common prediction task: phylogenetic tree reconstruction for a given collection of sequences (Abadi 2020).
A research study gave a detailed analysis of plant diversity trends to date, demonstrating that using machine learning to forecast future diversity could be tremendously beneficial. They applied machine learning approaches to phylogenetic diversity in vascular plants (Park 2020). Bhattacharjee et al., for the very first time, demonstrated the potential and feasibility of using deep learning techniques to compute distance matrices. The study evaluated both matrix factorization (ME) and autoencoder (AE) and aimed to develop improvised models for better results. They showed that both these methods are reliable and can be applied for handling large-scale datasets. They also highlighted the ability of these techniques over the heuristic-based techniques to automatically learn complicated inter-variable associations. Their research can also be used as a model for applying machine learning methods to the phylogenetic analysis (Bhattacharjee 2020).
In another research, a machine learning framework was developed to rank the neighbouring trees in accordance with their prosperity to increase the likelihood. They applied multiple features and utilized machine learning to improve an optimal tool. The study suggested specific ways to practice machine learning algorithms in phylogenetic analysis. Furthermore, they presented a methodology that can significantly speed up tree-search algorithms without sacrificing accuracy(Azouri 2021).
A recent review focused on the application of machine learning-based techniques in the data analysis of the human microbiome. It provided an insight into the plethora of advantages that machine learning has to offer over classical methods. The most common techniques covered in this review involved Support Vector Machines, Random Forest, k-NN and Logistic Regression. This review suggested how machine learning can contribute to the development of new models that can be useful in predicting classifications in the field of microbiology, inferring host phenotypes to predict diseases and characterization of state-specific microbial signatures using microbial communities(Macros 2021).
Future scope:
All the recently conducted research emphasizes the potential of artificial intelligence and machine learning in the inference of phylogenetic trees. These studies highlight the ability of machine learning in elevating the scale of analyzed datasets and the degree of sophistication in evolutionary models(Azouri 2021). Machine learning can thus be of high interest in the near future and contribute to efficient phylogenetic analysis in biological research.
| METHODS | DOMAIN | PURPOSE | References |
| Machine learning & Phylogenetic analysis | TSS (The Substitution Score) ISS (Internal substitution score) | To predict the pathogenecity of human mtDNA variants | (Akpinar 2020). |
| Machine learning & Phylogenetic analysis | ModelTeller (computational methodology) | To examine the accuracy of phylogenetic analyses, using machine learning | (Abadi 2020). |
| Machine learning & Phylogenetic analysis | Random Forest (RF) based learning and NeoPLE (prediction approach) | To highlight the use of candidate trees and successfully establish a model that can describe the relationship between likelihood and extracted features through the exploitation of deep neighbor information of each individual tree | (Ling 2020) |
References
- Ling, C., Cheng, W., Zhang, H., Zhu, H., & Zhang, H. (2020). Deep Neighbor Information Learning From Evolution Trees for Phylogenetic Likelihood Estimates. IEEE Access, 8, 220692-220702.
- Azouri, D., Abadi, S., Mansour, Y., Mayrose, I., & Pupko, T. (2021). Harnessing machine learning to guide phylogenetic-tree search algorithms. Nature communications, 12(1), 1-9.
- Abadi, S., Avram, O., Rosset, S., Pupko, T., & Mayrose, I. (2020). ModelTeller: model selection for optimal phylogenetic reconstruction using machine learning. Molecular Biology and Evolution, 37(11), 3338-3352.
- Park, D. S., Willis, C. G., Xi, Z., Kartesz, J. T., Davis, C. C., & Worthington, S. (2020). Machine learning predicts large scale declines in native plant phylogenetic diversity. New Phytologist, 227(5), 1544-1556.
- Akpinar, B. A., Carlson, P. O., Paavilainen, V. O., & Dunn, C. D. (2020). Pathogenicity of human mtDNA variants is revealed by combining a novel phylogenetic analysis with machine learning. bioRxiv.

 Previous Post
Previous Post Next Post
Next Post